Each type of data (Pathway, Reaction, Enzyme, and Compound) in PMN databases has a detail page which can be accessed by the various search functions or through hyperlinks from other data detail pages. This section describes the Pathway Detail, Reaction Detail, Enzyme Detail, and Compound Detail pages.
Each pathway is represented on detail page in a zoomable graphical format along with links to information about the reactions, reactant and product compounds, enzymes and genes.
Pathway Diagram: Each pathway contains some of the following color-coded information.
Zoom Level Controls: The upper portion of the pathway detail page has two buttons which allow you to zoom in (or out) on the diagram.
Evidence: Each pathway is associated with an evidence code which indicates the type of data used to support the existence of the pathway. For more specific information, see the tutorial on evidence codes.
Superclasses: Pathways are classified into a hierarchy of superclasses.
Species Data Available for: It is assumed that the entire pathway presented in the diagram is present in all of the species listed.
Summary: Pathway detail page include a brief summary composed by the database curators.
References: For pathways that have been curated, the literature used as references are hyperlinked in this section.
Unification Links: This section contains links to other databases containing related information.
Pathway Evidence Glyph: The pathway evidence glyph is displayed only for pathways that have been computationally predicted.
The reaction detail pages can be accessed directly from the search results (e.g. when searching for a specific reaction) and from the list browser. They are also linked from related pathway, compound and enzyme detail pages.
Superclasses: The reaction hierarchy represented in PMN databases is derived from the Enzyme Commission (EC) classification.
Pathways: Each reaction detail page lists all of the pathways in which the reaction is known or predicted to occur.
Reaction Diagram: A diagram of the reaction is displayed in accordance with the direction of that reaction within a pathway where the substrate compounds are shown to the left of the equation and the product compounds to the right.
Gene-Reaction Schematic: The gene-reaction schematic is a graphical representation of the relationship among sets of genes, enzymes and reactions.
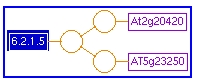
Unification Links: As with the pathway detail page, the unification links point to other databases that describe enzymes, reactions and proteins in the reaction.
The Enzyme detail pages can be accessed directly from the search results, from the list browser and from other detail pages.
Synonyms: Alternative names for the enzyme are displayed.
Superclassess: As with the pathways, reactions and compounds, enzymes are ordered into a hierarchy which can be browsed from this section.
Comments: Comments include anything that the curators determine are useful information about the enzyme such as specific information about the isozymes, physico-chemical properties (e.g. pH, temperature optimum) and kinetic properties (e.g. Km).
Species: The species indicates the species(es) in which this enzyme is found.
Gene: The gene(s) that encode for the protein(s) that make up the enzyme are listed.
If an enzyme is multimeric and comprised of different protein products, all of the genes encoding the proteins will be listed here.
Gene-Reaction Schematic: This is the same diagram that is displayed on the reaction detail page.
In the diagram, the enzyme being displayed is indicated by the filled yellow circle.
Enzymatic reaction: The reaction(s) catalyzed by the enzyme are displayed, with the equation set up to reflect the observed directionality.
Detailed information about substrate and product compounds can be accessed directly from the database search, or via links from other data detail pages.
Synonyms: Each compound may have one or more synonyms which are alternative names for the compound.
Superclasses: Compounds are grouped into classes, which indicate their relationship to other classes of compounds.
Empirical formula and molecular weight: Basic information about the compound such as its empirical formula and molecular weight are shown here. This information can be useful for analyzing metabolomic data.
Structure: The structure of the compound is shown.
SMILES Profile: SMILES is widely used as a general-purpose chemical nomenclature and data exchange format.
Unification Links: As with the other detail pages, links to similar or related information about the compound are listed here and hyperlinked to the corresponding database entry.
Participation in reactions
Please contact us if you have any questions or check our FAQs page for answers to common questions.
In order to assess quality of the data in the PMN databases, evidence codes are associated to the pathways and enzymes.
In the case of computationally derived pathways, color coding of the pathway glyph indicates how well supported individual reactions are.
Evidence Codes
All the databases in PMN use a controlled vocabulary of evidence codes. The three main classes (experimental, author statement and computational) of evidence are graphically represented on database detail pages.
The evidence codes are organized into an ontology that is available from the SRI Pathway Tools site.
An icon representing the type of evidence used to support the pathway is shown in the upper right hand corner of each pathway detail page. There are four kinds of icons:
As an example, take a look at the pathway detail page for dTDP-L-rhamnose biosynthesis II in AraCyc.
Evidence codes for enzymatic activities
Enzymes catalyzing reactions are also annotated with evidence codes. The same codes and icons are used for enzymes and pathways.
Using the same example pathway as before, GABA degradation, zoom in on the pathway using the 'more detail' button.
Understanding the pathway evidence glyphs
The pathway evidence glyph on the pathway detail page for computationally-predicted pathways is another source of information about how well supported any pathway is.
Note: Pathway glyphs are NOT present for pathways that have experimental, literature-based, or curator support.
In the pathway evidence glyph:
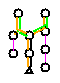
An example of a pathway evidence glyph is seen in the bottom of the Beta-Alanine Biosynthesis I detail page in AraCyc.
The pathway evidence glyph helps to determine whether a computationally-predicted pathway should be retained in the database.
The pathway evidence glyph helps to identify so-called "pathway holes" where the enzymes which catalyze specific reactions have not yet been identified.
Please contact us if you have any questions or check our FAQs page for answers to common questions.
Please help us improve the PMN
We are grateful for any data or other information that users are willing to submit to the PMN, either to expand our coverage or to correct mistakes in our existing entries.
*Please note that incomplete information is also welcome; ANY partial information you can give us will help us to make the PMN a better resource for our users.
But, we request that you send references whenever possible.
There are four main options for contributing information
Good file formats:
If you don’t see your file type here, please send it anyway; we will let you know if we can’t open it.
Filling in Data Submission / Correction Forms
Please keep in mind the following while filling out the forms:
*After you enter the detailed information for a reference once, please use the (Author Year) convention to refer back to the same reference, using a, b, etc., if necessary.
Columns A-H ask for basic information about the compound
Columns J-M ask for more detailed information
Please give a common or scientific name for the compound that should be used as the primary name in a database entry.
*Please use the EXACT compound name found in our databases if you are suggesting a correction.
Please select the appropriate term using the menu that is displayed when you click on the triangle to the right side of the cell.
Please give as many names as you feel are appropriate or would be helpful. Please keep in mind that the program that allows overlays of metabolic data will be more successful if it has more synonyms to match.
We appreciate any and all IDs that we can use to link our compounds to other databases. Common ID sources include: PubChem, KEGG, CAS, ChEBI, but please send us any other type of ID as long as you inform us of the source.
We do not currently use a formal chemical ontology to classify our compounds, so please give us any classification term that you use. You can assign more than one term to a compound, such as "secondary metabolite" and "terpenoid". The ChEBI site offers a good ontology if you need any suggestions.
If you have additional references that have not been included in other columns, please provide them here. We welcome references for published data, but we will also gratefully accept links to lab websites, on-line databases, etc, where we can get more information about this compound.
See good reference formats above.
Please enter any free text you would like about the compound, including its interesting properties, biological relevance in plants, benefit (or harm) to humans and other organisms, etc. Please include any references you have that support the comments.
More Detailed Compound Information
We welcome any information about the part(s) of the cell where your compound is made or stored. Please include any references you have that support the assignments.
We welcome any information about the part(s) of the plant where your compound is made or stored or transported through. Please include any references you have that support the assignments.
Do you have the SMILES code for your compound on hand? If so, please just paste it in and we can readily create a structure.
If you have some piece of information that doesn't seem to fit anywhere else on the form, (e.g. an e-mail address for an expert in the field, a notification that this compound will show up in a pathway that you plan to submit soon, a personal greeting to the curator who might be working on the submission, etc.), please put it here.
Columns A-I ask for basic information about the enzyme
Columns J-Z ask for more detailed information
Basic Enzyme / Reaction Information
Please give us an enzyme name that describes its specific biochemical activty. We realize that it may be hard to include specific reactants or products in the enzyme name, but please do whenever possible.
Good names:
Bad names:
*Please use the EXACT enzyme name found in our databases if you are suggesting a correction.
Please select the appropriate term using the menu that is displayed when you click on the triangle to the right side of the cell.
Please give as many names as you feel are appropriate or would be helpful. In this column we accept both "good" and "bad names." Please keep in mind that the Pathway tools program that the PMN uses to predict the metabolic pathways in new species will benefit from having additional names entered as synonyms.
If you have additional references that have not been included in other columns, please provide them here. We welcome references for published data, but we will also gratefully accept links to lab websites, on-line databases, etc, where we can get more information about this enzyme and/or reaction.
See good reference formats above.
If this reaction is part of one or more biochemical pathways, please enter this information. We welcome short answers such as "glycolysis" or "ornithine biosynthesis" but you can also submit a text description, such as, "an auxin conjugation reaction that generates an intermediate that can be stored or catabolized via further oxidation."
Please enter any free text you would like about the enzyme or reaction, including interesting properties, biological relevance in plants, the benefit (or harm) of the products to humans and other organisms, etc. Please include any references you have that support the comments.
More Detailed Enzyme / Reaction Information
We welcome any information about the part(s) of the cell where your enzyme is located or your reaction occurs. Please include any references you have that support the assignments.
We welcome any information about the part(s) of the plant where your enzyme acts and/or is made or stored, and/or where the reaction occurs. Please include any references you have that support the assignments.
Please give the weight in kD and indicate whether the weight is experimentally determined (with a reference) or calculated based on the protein sequence.
If your enzyme is part of a complex, please describe both the identity and number of the subunits in the complex (with a reference). If there is only ambiguous data, e.g. that that enzyme A and B form a complex but the stoichiometry is not known, please give us that information, too.
Please provide any information about substances known to activate the enzyme (with a reference). If you know whether the activator is physiologically relevant, and the mode of activation, please give us that information, too.
Please provide any information about substances known to inhibit the enzyme (with a reference). If you know whether the inhibitor is physiologically relevant, and the mode of inhibition (e.g. allosteric, competitive), please give us that information, too.
Please provide any information known about cofactors or prosthetic groups that are required for enzyme activity (with a reference).
We welcome data concerning the Km of the enzyme. Please be sure to include the exact substrate used to measure the Km and include the units of measurement (with a reference). If the experiment was conducted at a particular pH, temperature, etc., this can be noted as well.
We welcome data concerning the Temperature optimum of the enzyme. Please be sure to include the exact substrate used to measure the T(opt) (with a reference). If the experiment was conducted at a particular pH, this can be noted as well.
We welcome data concerning the pH optimum of the enzyme. Please be sure to include the exact substrate used to measure the pH(opt) (with a reference). If the experiment was conducted at a particular temperature, this can be noted as well.
If you HAVE NOT entered an accession number of any kind in column H (Gene / Protein sequence identifier or name), but you do have access to the protein sequence, please paste it here. We welcome any additional data you can give. For example, is this protein prediction based on a predited gene model or is it based on an experimentally verified full-length cDNA sequence?
If you have some piece of information that doesn't seem to fit anywhere else on the form, (e.g. an e-mail address for an expert in the field, a notification that this reaction will show up in a pathway that you plan to submit soon, a comment on the importance of the PMN to your work, etc.), please put it here.
*Please note that on this form, each step in the pathway should be entered as a separate row.
- You may enter multiple pathways on the same page, or you may use the additional duplicate sheets provided to enter each distinct pathway. Please let us know if you have submitted more than one pathway in your e-mail message.
- Since some of the information might be the same for each step of the pathway (e.g.Pathway synonym(s)), or for several steps of the pathway (e.g. sub-cellular localization). You are welcome to copy and paste the duplicated information or you can use the notation: "same as row X" where X is the number of the row where the duplicate data first appeared.
If you are submitting a correction to a pathway, you do not need to resubmit the parts of the pathway that are correct. Please just enter the new information that should replace the erroneous step(s).
Please generate a name for the pathway. A good name typically includes the name of a final product, or class of products generated, and a descriptive word, such as "biosynthesis" or "degradation" to indicate the biological purpose of the pathway.
Please select the appropriate term using the menu that is displayed when you click on the triangle to the right side of the cell.
Please give any suggested synonyms for the pathway. For example, "auxin biosynthesis" could be a synonym for "IAA biosynthesis" that might be helpful to future users performing searches.
Please list a name for the enzyme or enzymes that carry out the reaction and provide a reference if possible. You can add additional synonyms using a separate enzyme form.
*A gene or protein ID (from NCBI, TAIR, UniProt, etc.) for the enzyme would be very helpful.
If you have additional references that have not been included in other columns, please provide them here. We welcome references for published data, but we will also gratefully accept links to lab websites, on-line databases, etc, where we can get more information about this pathway, enzyme and/or reaction.
See good reference formats above.
We welcome any information about the part(s) of the cell where each step of the pathway occurs. Please include any references you have that support the assignments.
We welcome any information about the part(s) of the plant where the reaction occurs. Please include any references you have that support the assignments.
If you have some piece of information that doesn't seem to fit anywhere else on the form, (e.g. an e-mail address for an expert in the field, a notification that you plan to submit a related pathway found in other organisms soon, an invitation for PMN members to attend an upcoming meeting you're hosting, etc.), please put it here.
Please contact us if you have any questions or check our FAQs page for answers to common questions.