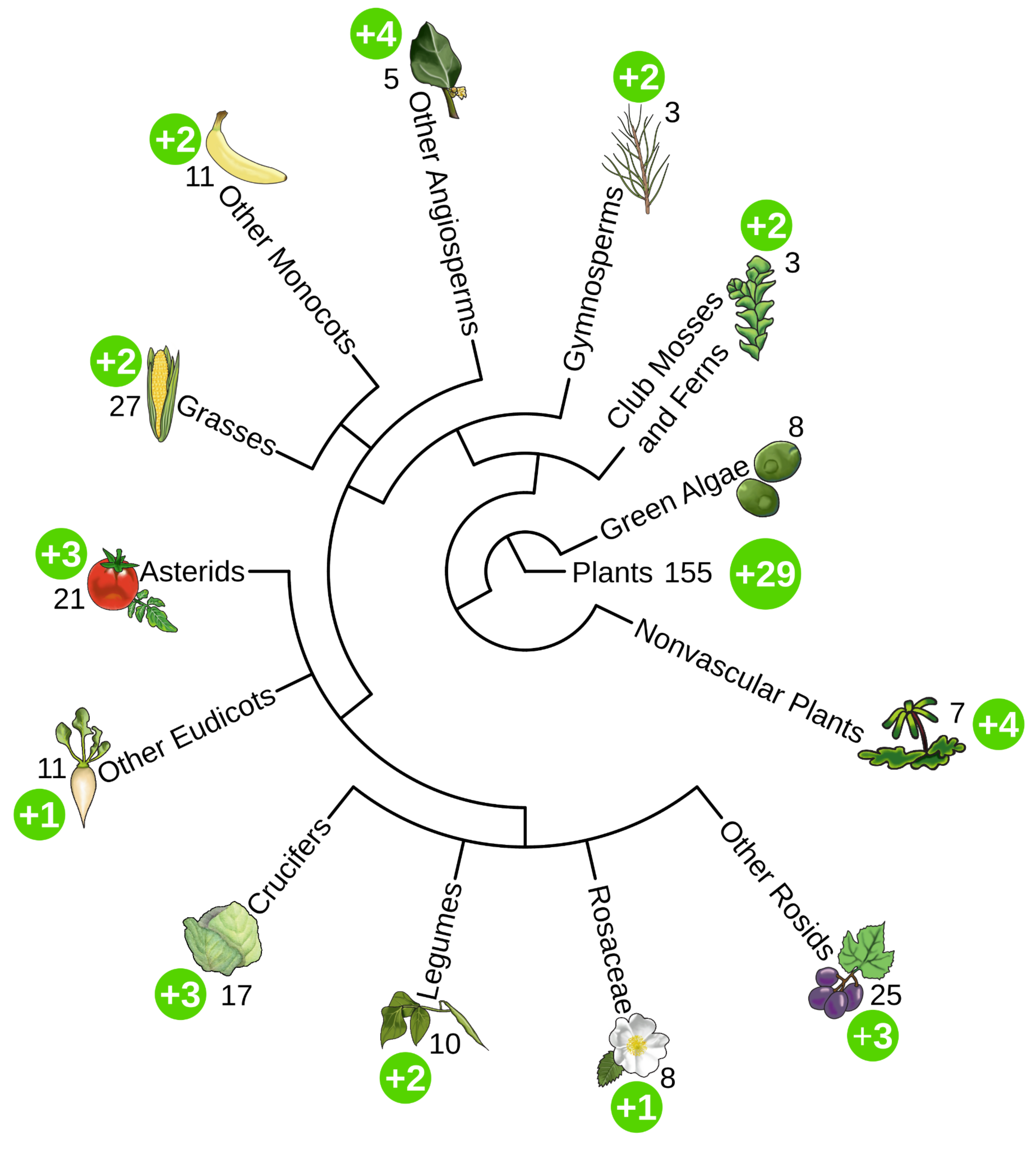
PMN 17 Released
We are super excited to announce the release of PMN 17! PMN 17 is the largest release in the history of PMN. Enabled by our new backend pipeline, PMN 17 includes 583 single-species databases, including updated versions of the 155 databases from PMN 16